A lexical approach for identifying behavioral action sequences was published in PLOS Computational Biology
- January 11, 2022
- Neural Information
Title: A lexical approach for identifying behavioral action sequences [PLOS Computational Biology / bioRxiv]
Authors: Gautam Reddy, Laura Desban, Hidenori Tanaka, Julian Roussel, Olivier Mirat, Claire Wyart
Published on 10 January 2022.
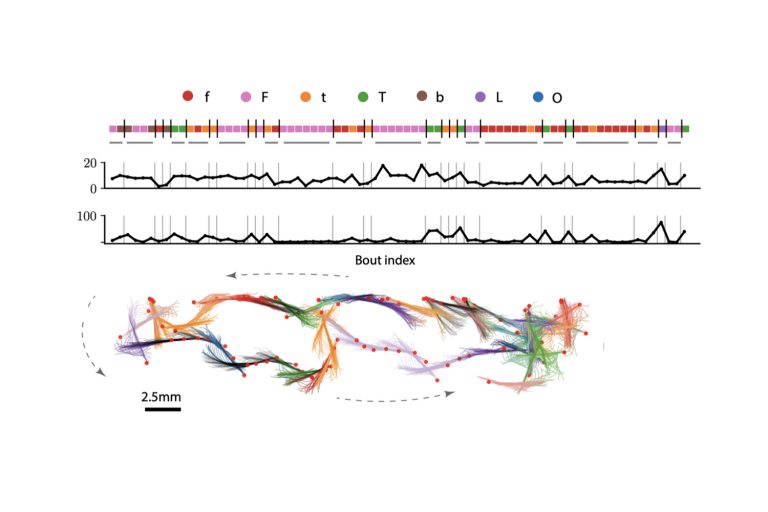
Abstract: Animals display characteristic behavioral patterns when performing a task, such as the spiraling of a soaring bird or the surge-and-cast of a male moth searching for a female. Identifying such conserved patterns occurring rarely in noisy behavioral data is key to understanding the behavioral response to a distributed stimulus in unrestrained animals. Existing models seek to describe the dynamics of behavior or segment individual locomotor episodes rather than to identify occasional, transient irregularities that make up the behavioral response. To fill this gap, we develop a lexical, hierarchical model of behavior. We designed an unsupervised algorithm called “BASS” to efficiently identify and segment conserved behavioral action sequences transiently occurring in long behavioral recordings. When applied to navigating larval zebrafish, BASS extracts a dictionary of remarkably long, non-Markovian sequences consisting of repeats and mixtures of slow forward and turn bouts. Applied to a novel chemotaxis assay, BASS uncovers conserved chemotactic strategies deployed by zebrafish to avoid aversive cues consisting of sequences of fast large-angle turns and burst swims. In a simulated dataset of soaring gliders climbing thermals, BASS finds the spiralling patterns characteristic of soaring behavior. In both cases, BASS succeeds in identifying action sequences that are highly conserved but transient in the behavior deployed by freely moving animals. BASS can be easily incorporated into the pipelines of existing behavioral analyses across diverse species, and even more broadly used as a generic algorithm for pattern recognition in low-dimensional sequential data.